HIF1A Antibodies
Background
HIF1A as a key transcriptional regulation factor, mainly exist in the cytoplasm of mammals. This gene is activated under hypoxic conditions and promotes the maintenance of oxygen homeostasis and metabolic adaptation by regulating the expression of downstream target genes, ensuring the survival of cells in hypoxic environments. Tumor cells often utilize the HIF1A pathway to respond to fluctuations in oxygen concentration in the tumor microenvironment. This gene was discovered by the Semenza team in 1992, and its protein structure was resolved in 2001, revealing a unique DNA binding mechanism mediated by the bHLH-PAS domain. HIF1A, as a milestone in oxygen sensing research, has greatly promoted the exploration of mechanisms and the development of targeted drugs in the fields of hypoxia signal transduction, cancer treatment, and ischemic diseases.
Structure of HIF1A
HIF1A is a protein with a molecular weight of approximately 93 kDa. Its precise molecular weight varies slightly among different species due to differences in amino acid composition.
| Species | Human | Mouse | Rat | Rhesus monkey | Pig |
| Molecular Weight (kDa) | 92.7 | 92.9 | 92.8 | 92.6 | 92.9 |
| Primary Structural Differences | Contains the bHLH-PAS domain | ODDD domain structure has two amino acid differences | PAS - B highly conserved structure domain | CTAD domain structure is similar to human height | Individual amino acid substitutions are present in the bHLH domain |
HIF1A is composed of approximately 826 amino acids and adopts a characteristic three-dimensional folded conformation of helical-ring-helical. This protein structure contains the key BHLH-PAS domain, where bHLH mediates DNA binding and the PAS domain is responsible for protein dimerization. There exists an oxygen-dependent degradation domain (ODDD) at the molecular center, which contains the prolyl hydroxylation site of conserved. The hydroxylation state of this site directly regulates the stability of the protein. The C-terminal trans activation domain (CTAD) initiates the expression of hypoxia response genes by interacting with transcriptional co-activators. The stability of the entire protein is precisely regulated by oxygen concentration: it is degraded through the ubiquitination pathway mediated by VHL under normoxic conditions, and forms a stable heterodimer in a hypoxic environment, which then enters the cell nucleus to regulate gene transcription.
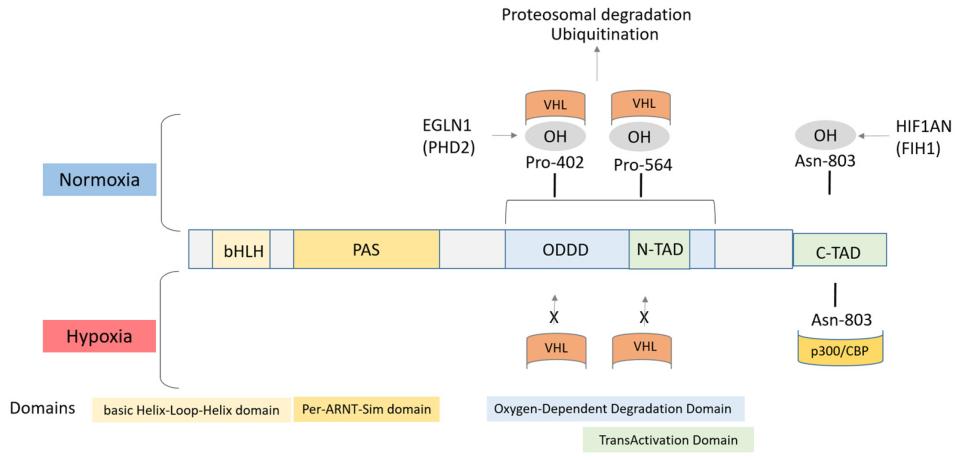 Fig. 1 Structure of the HIF1α protein. 1
Fig. 1 Structure of the HIF1α protein. 1
Key structural properties of HIF1A:
- Characteristic bHLH-PAS domains mediate DNA binding and dimerization
- Oxygen-dependent degradation domain (ODDD) including conservative preserved ammonia acyl hydroxyl loci
- The C-terminal trans activation domain (CTAD) recruits transcriptional co-activation complexes
- Oxygen sensing mechanisms regulate protein stability via prolyl hydroxylase
- Nuclear localization signals mediate nuclear and cytoplasmic shuttling under hypoxic conditions
Functions of HIF1A
The core function of HIF1A is to mediate the adaptive response of cells to hypoxic environments. In addition, it is widely involved in a variety of important physiological and pathological processes such as metabolic reprogramming, angiogenesis and cell survival.
| Function | Description |
| Regulation of hypoxia response | The expression is stable under hypoxic conditions, and the transcription of a variety of target genes is initiated after being transferred into the nucleus to promote cell adaptation to hypoxic microenvironment. |
| Metabolic reprogramming | Up-regulate the expression of glycolycle-related genes, inhibit oxidative phosphorylation, and shift cells to anaerobic metabolism during hypoxia to maintain energy supply. |
| Promotion of angiogenesis | Induce the expression of genes such as vascular endothelial growth factor (VEGF), promote the formation of new blood vessels, and improve tissue oxygen supply. |
| Inhibition of apoptosis | Regulating apoptosis-related genes and enhancing the survival ability of cells under hypoxic pressure is particularly significant in tumor cells. |
| Regulation of erythropoiesis | Promote the expression of erythropoietin (EPO), stimulate red blood cell production, and enhance the blood's oxygen-carrying capacity. |
The activity of HIF1A is precisely regulated by oxygen concentration: it rapidly degrades through the ubiquitination pathway under normoxic conditions, with a half-life of less than 5 minutes. However, in a hypoxic environment, it rapidly accumulates and activates, and its dose-effect curve shows a steep S shape, indicating that cells achieve extremely high sensitivity and rapid response ability to changes in oxygen concentration through this protein.
Applications of HIF1A and HIF1A Antibody in Literature
1. Yang, Kaixin, et al. "Long non-coding RNA HIF1A-As2 and MYC form a double-positive feedback loop to promote cell proliferation and metastasis in KRAS-driven non-small cell lung cancer." Cell Death & Differentiation 30.6 (2023): 1533-1549. https://doi.org/10.1038/s41418-023-01160-x
Research has found that in non-small cell lung cancer, KRAS can recruit DHX9 protein through MYC-induced lncRNA HIF1A-AS2, epigenetically activate MYC transcription, form a positive feedback loop, and promote tumor proliferation and metastasis. Targeted inhibition of HIF1A-AS2 can enhance the sensitivity to MYC inhibitors and chemotherapy.
2. Pelucchi, Sara, et al. "Hif1a: A putative modifier of hemochromatosis." International Journal of Molecular Sciences 22.3 (2021): 1245. https://doi.org/10.3390/ijms22031245
Research has found that the p.Phe582Ser and p.Ala588Thr variations of the HIF1A gene may serve as expression modifiers for HFE-related hereditary hemochromatosis (HH). These variations down-regulate hepcidin synthesis, up-regulate target genes such as VEGF, exacerbate iron overload and clinical phenotypes, and reveal the severe genetic complexity of HFE-HH.
3. Kunej, Tanja. "Integrative map of HIF1A regulatory elements and variations." Genes 12.10 (2021): 1526. https://doi.org/10.3390/genes12101526
This study constructed an integrated regulatory map of the HIF1A gene, covering its transcription, post-transcriptional regulatory factors, downstream target genes and genetic variations. Through multi-omics analysis, four harmful missense variations and 85 targeted mirnas were identified, revealing the complex network of HIF1A in hypoxia response and providing a systematic perspective for studying its role in diseases.
4. Liang, Zongwen, et al. "Elevated Histone Lactylation Mediates Ferroptosis Resistance in Endometriosis Through the METTL3‐Regulated HIF1A/HMOX1 Signaling Pathway." Advanced Science (2025): e08220. https://doi.org/10.1002/advs.202408220
Research has found that histone lactation modification in endometriosis mediates ferroptosis resistance through the HIF1A/HMOX1 signaling pathway regulated by METTL3. Targeted inhibition of lactation and induction of ferroptosis (such as 2-DG combined with erastin) can provide a new strategy for the treatment of this disease.
5. Brandt, Camilla Blunk, et al. "HIF1A Knockout by Biallelic and selection-free CRISPR gene editing in human primary endothelial cells with ribonucleoprotein complexes." Biomolecules 13.1 (2022): 23. https://doi.org/10.3390/biom13010023
This study established an efficient and non-selective CRISPR gene editing method based on RNP, successfully achieving biallelic knockout of the HIF1A gene in primary human umbilical vein endothelial cells (HUVECs) with an efficiency of 98%, and verified the effects of HIF1A deletion on angiogenesis and hypoxia response, providing a reliable technical means for the study of endothelial cell function.
Creative Biolabs: HIF1A Antibodies for Research
Creative Biolabs specializes in the production of high-quality HIF1A antibodies for research and industrial applications. Our portfolio includes monoclonal antibodies tailored for ELISA, Flow Cytometry, Western blot, immunohistochemistry, and other diagnostic methodologies.
- Custom HIF1A Antibody Development: Tailor-made solutions to meet specific research requirements.
- Bulk Production: Large-scale antibody manufacturing for industry partners.
- Technical Support: Expert consultation for protocol optimization and troubleshooting.
- Aliquoting Services: Conveniently sized aliquots for long-term storage and consistent experimental outcomes.
For more details on our HIF1A antibodies, custom preparations, or technical support, contact us at email.
Reference
- Kunej, Tanja. "Integrative map of HIF1A regulatory elements and variations." Genes 12.10 (2021): 1526. https://doi.org/10.3390/genes12101526
Anti-HIF1A antibodies
 Loading...
Loading...
Hot products 
-
Mouse Anti-C1QC Recombinant Antibody (CBFYC-0600) (CBMAB-C0654-FY)

-
Rabbit Anti-AKT3 Recombinant Antibody (V2-12567) (CBMAB-1057-CN)

-
Mouse Anti-CASP8 Recombinant Antibody (CBYY-C0987) (CBMAB-C2424-YY)

-
Mouse Anti-AHCYL1 Recombinant Antibody (V2-180270) (CBMAB-A1703-YC)

-
Mouse Anti-CD247 Recombinant Antibody (6B10.2) (CBMAB-C1583-YY)

-
Mouse Anti-APP Recombinant Antibody (5C2A1) (CBMAB-A3314-YC)

-
Mouse Anti-CFL1 (Phospho-Ser3) Recombinant Antibody (CBFYC-1770) (CBMAB-C1832-FY)

-
Mouse Anti-CD46 Recombinant Antibody (CBFYC-0076) (CBMAB-C0085-FY)

-
Mouse Anti-dsDNA Recombinant Antibody (22) (CBMAB-AP1954LY)

-
Mouse Anti-ARHGDIA Recombinant Antibody (CBCNA-009) (CBMAB-R0415-CN)

-
Mouse Anti-ALB Recombinant Antibody (V2-180650) (CBMAB-A2186-YC)

-
Mouse Anti-BCL6 Recombinant Antibody (CBYY-0435) (CBMAB-0437-YY)

-
Mouse Anti-ACKR3 Recombinant Antibody (V2-261265) (CBMAB-C1023-LY)

-
Rat Anti-CCR2 Recombinant Antibody (475301) (CBMAB-C1338-LY)

-
Mouse Anti-CCS Recombinant Antibody (CBFYC-1093) (CBMAB-C1150-FY)

-
Rabbit Anti-ALOX5AP Recombinant Antibody (CBXF-1219) (CBMAB-F0750-CQ)

-
Mouse Anti-CD8 Recombinant Antibody (C1083) (CBMAB-C1083-LY)

-
Mouse Anti-FOXA3 Recombinant Antibody (2A9) (CBMAB-0377-YC)

-
Mouse Anti-ACE2 Recombinant Antibody (V2-179293) (CBMAB-A0566-YC)

-
Mouse Anti-BRCA2 Recombinant Antibody (CBYY-1728) (CBMAB-2077-YY)

- AActivation
- AGAgonist
- APApoptosis
- BBlocking
- BABioassay
- BIBioimaging
- CImmunohistochemistry-Frozen Sections
- CIChromatin Immunoprecipitation
- CTCytotoxicity
- CSCostimulation
- DDepletion
- DBDot Blot
- EELISA
- ECELISA(Cap)
- EDELISA(Det)
- ESELISpot
- EMElectron Microscopy
- FFlow Cytometry
- FNFunction Assay
- GSGel Supershift
- IInhibition
- IAEnzyme Immunoassay
- ICImmunocytochemistry
- IDImmunodiffusion
- IEImmunoelectrophoresis
- IFImmunofluorescence
- IGImmunochromatography
- IHImmunohistochemistry
- IMImmunomicroscopy
- IOImmunoassay
- IPImmunoprecipitation
- ISIntracellular Staining for Flow Cytometry
- LALuminex Assay
- LFLateral Flow Immunoassay
- MMicroarray
- MCMass Cytometry/CyTOF
- MDMeDIP
- MSElectrophoretic Mobility Shift Assay
- NNeutralization
- PImmunohistologyp-Paraffin Sections
- PAPeptide Array
- PEPeptide ELISA
- PLProximity Ligation Assay
- RRadioimmunoassay
- SStimulation
- SESandwich ELISA
- SHIn situ hybridization
- TCTissue Culture
- WBWestern Blot








